Scattering
Small Angle X-Ray and Neutron Scattering (SAXS/SANS)
Equipment, Experiments and Modelling
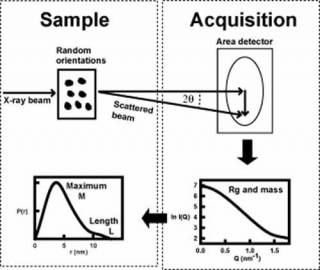
Scattering experiments study the overall structure of macromolecules in random orientations in solution1. This is a diffraction experiment akin to crystallography, but in a scattering experiment, the crystal is replaced by molecules free in solution. The intensities of the scattering curve are measured as a function of scattering angle. From this, the radius of gyration Rg (degree of elongation) and molecular mass are determined, together with dimensional information on macromolecular shape and length L. Thus scattering provides a multi-parameter description of a protein structure in near-physiological conditions, but at low structural resolution (2-4 nm). This method is especially useful for macromolecules that have not been crystallised.
We perform our scattering work on the X-ray instruments B21 at the Diamond Light Source synchrotron and on ID02 and BM29 at the European Synchrotron Radiation Facility and on the neutron instruments SANS2D at the ISIS neutron spallation source and D22 at the Institut Laue-Langevin neutron reactor.
Atomistic modelling based on known crystal structures is able to extract further structural information from the scattering curves in molecular details. This modelling is based on randomizing arrangements of any known subunits that are joined by flexible linkers via Monte Carlo simulations (eg: the hinges in antibody molecules), or by applying molecular dynamics simulations to the whole macromolecule (eg: collagen triple helices). The large libraries of physically-realistic structures are compared with the scattering data to identify the best fit structures. The resulting molecular structures can provide new insights into structures and functions of these biological macromolecules. The best-fit scattering structures are made publicly available.
Publications:
Perkins, S. J. (2009). Unravelling antibody and complement structures in immunity using TS-2 neutrons at ISIS: Neutron scattering. The Biochemist. 31 (4), 32-35.
Perkins, S. J., Wright, D. W., Zhang, H., Brookes, E. H., Chen, J., Irving, T. C., Krueger, S., Barlow, D. J., Edler, K. J., Scott, D. J., Terrill, N. J., King, S. M., Butler, P. D. & Curtis, J. E. (2016) Atomistic modelling of scattering data in the Collaborative Computational Project for Small Angle Scattering (CCP-SAS). J. Appl. Crystall. 49, 1861-1875. doi:10.1107/S16005767160151X. Pubmed 27980506
Molecular IgG3 structure paves the way for new applications of antibodies
In humans, immunoglobulin (IgG) is the most common type of antibody found in blood circulation. IgG molecules are created by plasma B cells, and there are four subclasses called IgG1 to IgG4. Of the four, IgG3 is the least understood. It has a uniquely long hinge region separating its Fab antigen-binding and Fc receptor-binding regions. The presence of this elongated hinge makes it challenging to perform structural studies, for example, with X-ray crystallography. Our recent work used a combination of methods to provide the first experimentally determined molecular structural model for a full-length IgG3 antibody.
Small angle X-ray scattering (SAXS) and analytical ultracentrifugation (AUC) showed that IgG3 is elongated compared to IgG1, IgG2 and IgG4. SAXS also showed that IgG3 has a more extended central hinge than IgG1 and IgG2 that links its three globular regions together. Using atomistic modelling based on computational molecular dynamics and Monte Carlo methods to fit the scattering curves, the SAXS and SANS data sets were successfully modelled as molecular structures. The results of this joint AUC-SAXS approach confirm that the IgG3 molecule has a semi-rigid long hinge with little disorder, and results in a wide separation between the globular Fab and Fc regions.

Spiteri VA et al. Solution structures of human myeloma IgG3 antibody reveal extended Fab and Fc regions relative to the other IgG subclasses. Journal of Biological Chemistry, 100995 (2021). DOI: 10.1016/j.jbc.2021.100995.
Journal Front Cover. The new IgG3 structure is shown in red, and IgG1, IgG2 and IgG4 are shown in green, yellow and orange.
Sugar-free antibodies - Atomistic scattering modelling of human IgG1 reveals conformational changes on glycan removal
Antibodies are an important biotherapeutic tool: they can be tailored to treating different diseases, and can direct toxic drugs to the area in the body where they are needed. The computational modelling of antibodies alongside Small Angle Neutron and X-ray Scattering (SANS and SAXS) data determined the effect of bound sugars on their structural behaviour. We looked at the most common class of human antibodies called IgG1. These antibodies are shaped like a ‘Y’, and act as a bridge: the top two regions, the Fab regions, interact with the foreign material, and the bottom, known as the Fc region and decorated with sugars, interacts with your immune system via immune receptors. We measured the antibodies with and without sugars on the Sans2D instrument at ISIS, the B21 instrument at Diamond Light Source, and analytical ultracentrifuges at UCL. Small differences were detected between the two forms.
Combining the experimental data with a detailed computational modelling study, we produced 100 best-fit structures for each of the two IgG1 forms, using SASSIE-web, an online web server which streamlines the atomistic modelling of SAS data. These best-fit models demonstrated conformational differences of the Fc region following glycan removal that accounted for changes in the scattering data, and that the glycans stabilised the structure of the Fc region in intact IgG1.
Our combination of analytical ultracentrifugation, small angle X-ray, neutron scattering, and the atomistic modelling of these data sets has now been demonstrated to be powerful enough in probing more subtle structural changes such as glycan removal in antibodies. This methodology can be extended to other systems.

Spiteri, V. A., Doutch, J., Rambo, R. P., Gor, J., Dalby, P. A. & Perkins, S. J. (2021) Solution structure of deglycosylated human IgG1 shows the role of CH2 glycans in its conformation. Biophysical J. 200, 1814-1834. https://doi.org/10.1016/j.bpj.2021.02.038
Journal Front Cover. The ribbon cartoon images of the 16 IgG1 structures on the cover of the May 4 issue of Biophysical Journal were selected at random from the 100 and 100 best-fit structures for glycosylated and deglycosylated IgG1
 Close
Close

