
Sequencing at UCL Genomics
UCL Genomics offers support at every stage of the sequencing pipeline from experimental design, in-depth sample quality control, library preparation, to sequencing and data analysis
1. Quality Control
Sample Quality Control is the first step of the sequencing pipeline and one of the crucial steps for a successful sequencing experiment. Our labs house Qubit fluorometers, a plate reader, an Agilent Bioanalyser Agilent Tapestation 4200 and a Nanodrop.

2. Library preparation
UCL Genomics offers a wide range of library preparation kits for a range of next-generation sequencing applications. Select an application below to see the range of kits currently used in our facility. We ahve autromated solutions for must of the these kits so we can help with very large as well as smaller projects. UCL Genomics is constantly updating and testing new kits on the sequencing market and we are happy to discuss testing new technologies with you.
- RNA-seq
We can offer RNA-sequencing on a variety of sample types, including degraded (FFPE) samples and/or very low input (internally validated as low as 10pg). Samples can either be poly-A selected for mRNA sequencing, or ribosomally depleted to capture non-adenylated transcripts. We can also offer globin depletion if working from whole blood RNA.
Standard RNA-seq solutions at UCL Genomics
Library prep kit Recommended Input amount KAPA RNA HyperPrep kit 1 – 100 ng KAPA mRNA HyperPrep kit 50 ng – 1 µg KAPA HyperPrep kit with RiboErase (HMR) 25 ng – 1 µg KAPA RNA HyperPrep kit with RiboErase (HMR) Globin 25 ng – 1 µg NEBNext Ultra II Directional RNA Library Prep kit for Illumina 5 ng – 1 µg Low input RNA-seq solutions at UCL Genomics
Library prep kit Recommended Input amount Clontech SMARTER low input RNA kit followed by Illumina Nextera XT DNA kit Micro RNA-seq solutions at UCL Genomics
Library prep kit Recommended Input amount NEBNext Multiplex Small RNA Qiagen QIAseq miRNA Library kit 1 ng – 500 ng - ChiP-seq
- UCL Genomics have experience with both standard ChIP libraries, as well as running optimised protocols for “Cut-and-Run” & “Cut-and-Tag” samples. Samples can be prepped from as little as 1ng of immuno-precipitated DNA.
Library prep kit Recommended Input amount NEBNext DNA Ultra II Library prep kit 1 – 100 ng - Targeted-seq
UCL Genomics support both off-the-shelf and custom panels from Nonacus, IDT and Agilent. Libraries can be prepped from both genomic, FFPE and cfDNA for enrichment. Use of molecular identifiers in the Nonacus, IDT & SureSelect XT HS2 assays are supported to allow for the highest sensitivity assay when sequencing at high depths.
Library prep kit Recommended Input amount Nonacus Cell3 Target 10ng-1µg Agilent SureSelect XT 200 ng 100 – 200 ng Agilent SureSelect XT 3 µg 1 – 3 µg Agilent SureSelect XT Low Input 10 – 200 ng Agilent SureSelect XT HS2 10 – 200 ng - Pathogen Sequencing
- Our pathogen sequencing service provision includes:• Library preparation of whole pathogen genomes and amplicons
• Library preparation for enrichment of pathogen genomes including from clinical or FFPE samples using our own in-house custom design capture probes against a variety of DNA and RNA viruses and bacteria
• SARS-CoV-2 sequencing by targeted enrichment (Agilent), or ARTIC amplicon sequencing (Illumina and Oxford Nanopore)
• 16s rRNA metagenomics
• RNA-Seq for bacterial transcriptome analysis or pathogen discovery
• DNA and RNA metagenomicsWhole Pathogen Genomes and Amplicons
Library prep kit Recommended Input amount NEBNext DNA Ultra II Library prep kit
(choice of mechanical or enzymatic shearing)1 – 100 ng Illumina DNA Prep 1 – 500 ng
Targeted enrichmentLibrary prep kit Recommended Input amount Agilent SureSelect XT Dependent on pathogen load 16s rRNA metagenomics
Library prep kit Recommended Input amount Illumina protocol (V3-V4) 12.5 ng RNA-seq
Library prep kit Recommended Input amount KAPA RNA HyperPrep kit 1 – 100 ng KAPA HyperPrep kit with RiboErase (HMR) 25 ng – 1 µg DNA/RNA metagenomics
Library prep kit Recommended Input amount NEBNext DNA Ultra II Library prep kit
(choice of mechanical or enzymatic shearing)1 – 100 ng KAPA RNA HyperPrep kit 1 – 100 ng KAPA HyperPrep kit with RiboErase (HMR) 25 ng – 1 µg Capture probes designed by the facility are shown in the table below. In addition, we have listed those pathogens for which we are able to provide antimicrobial or antiviral resistance testing either from amplicons or whole genomes. We are happy to collaborate with customers to design and optimise new capture probe sets for a pathogen that is not already held by the facility. Bait sets already designed include
Pathogen WGS Typing Resistance
Detection
Adenovirus (ADV) ✓ ✓ Cytomegalovirus (CMV) ✓ ✓ ✓ Enteroviruses ✓ ✓ Epstein-Barr virus (EBV) ✓ ✓ Hepatitis B virus (HBV) ✓ ✓ ✓ Hepatitis C virus (HCV) ✓ ✓ ✓ Herpes simplex virus (HSV) ✓ ✓ ✓ Human coronaviruses (inc. SARS-CoV-2) ✓ ✓ Human herpesvirus 6 (HHV6) ✓ ✓ Human Immunodeficiency virus 1 (HIV-1) ✓ ✓ ✓ Human Immunodeficiency virus 2 (HIV-2) ✓ Human papillomavirus (HPV) ✓ ✓ Influenza A (FluA) and Influenza B (FluB) ✓ ✓ ✓ Norovirus ✓ ✓ Varicella zoster virus (VZV) ✓ ✓ ✓ Mycobacterium tuberculosis (Mtb) ✓ ✓ ✓ Chlamydia trachomatis (Ct) ✓ Neisseria meningitidis ✓ ✓ Legionella pneumophilia ✓ Pan-Entero-with-Rhinovirus ✓ Pan-Respiratory ✓ ✓ Parainfluenza, metapneumovirus, RSV ✓ - DNA-seq
UCL Genomics can perform library prep from both high molecular weight and FFPE DNA samples for whole genome sequencing. We have experience performing both low and high depth sequencing on a variety of genomes (from bacteria to human genomes). If sufficient input is provided, we now offer PCR free prep for Illumina sequencing for increased genome coverage.
Library prep kit Recommended Input amount NEBNext DNA Ultra II Library prep kit
(choice of mechanical or enzymatic shearing)500 pg – 1 µg Illumina PCR free library prep kit 25 ng - 1 µg Illumina DNA Prep 1 - 500 ng - Single Cell Sequencing
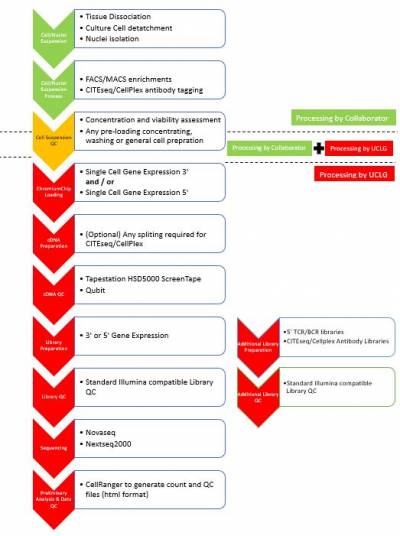
We are excited to be able to offer a single cell sequencing service at UCL Genomics, supported by GOSHCC. Using the 10X Genomics Chromium platform, UCL Genomics offers
- the standard Single Cell Gene Expression solution (3’ Gene Expression)
- Single Cell Immune Profiling (5’ Gene Expression with TCR/BCR sequencing).We also support projects utilising Feature Barcode Technology (e.g. CITESeq, CellPlex etc). We are accepting single cell/nuclei suspensions for processing from suspension QC and Chromium chip loading through sequencing and up to preliminary data analysis (generation of count matrix).
- Long read sequencing
UCL Genomics generate long read sequence using either the MinION or GridION instruments (Oxford Nanopre), depending on the amount of sequence data required for a particular project. The GridION platform allows us to run 5x MinION flow cells to achieve up to 250 Gbp data over the course of 48-72 hour, but this may vary according to library type and run conditions.
The main applications for ONT sequencing are:
• SARS-CoV-2 sequencing pipeline with ARTIC amplicons
• Whole Genome Sequencing
• RNA and cDNA Sequencing
• Targeting and enrichment strategies
• Metagenomics
3. Sequencing

UCL Genomics has a range of Illumina sequencers in-house and access to additional sequencers through our genomic partners. The facility also houses an Oxford Nanopore GridION and MinIONs.
4. Bioinformatics and software
UCL Genomics offers a tiered system for data analysis from standard pipelines for differential gene expression analysis to custom support for more in-depth analysis and analysis of pathogen sequencing data. The Facility hosts the Ingenuity Pathway Analysis (IPA) software which is available to use (for a charge)
 Close
Close


