All the images rendered using RasMol
Check here to download the dimer form of JNK1-alphal1 isoform of PDB and view PDB annotation at RCSB
Primary (citable) accession number: P45983
Crystal Structure of JNK1-alpha1 isoform
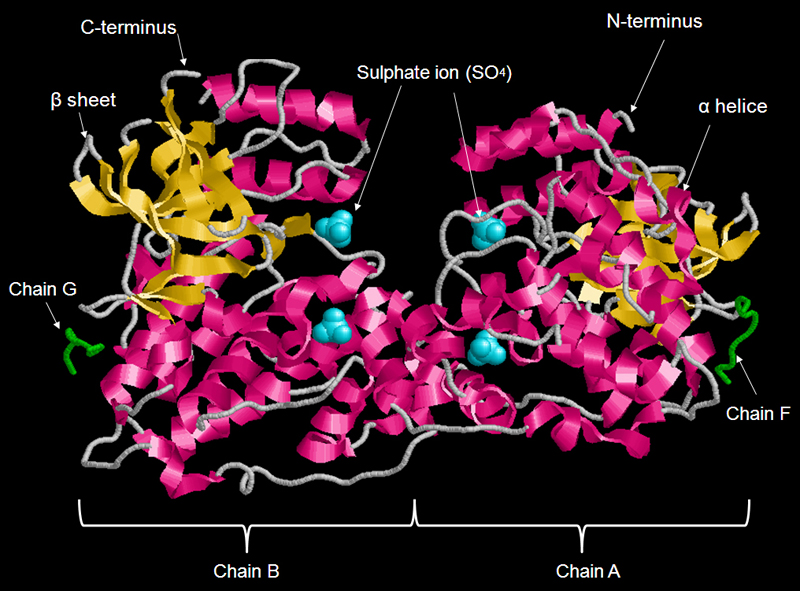
FIGURE 1 Structure of a dimer of JNK1 alpha-1 isoform with 4 polypeptide chains- chain A, B, G and F. JNK is regulated by a number of upstream MAPK kinases, which activate JNK by phosphorylation at residues 183 and 185 within the activation loop. Polypeptides A and B are compose for the mitogen-activated protein kinase 8 whereas polypeptides chain G and F are compose for C-Jun-amino-terminal kinase-interacting protein 1. Monomeric form of JNK1-alpha1 composes of two polypeptides (chain A/ B and chain F/G). Each JNK1 alpha-1 molecule have a number of different regions that share the features of all protein tyrosine kinase: protein kinase domain, active site, activation loop, ATP binding loop and KIM docking site.
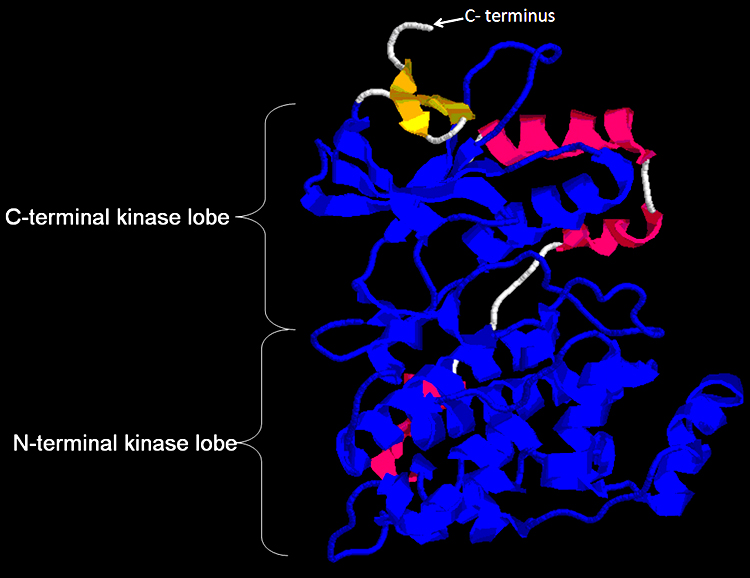
FIGURE 2 Crystal sturucture of the unphosphorylated form of protein kinase domain shows in blue.The protein kinase domain consists of amino acids 26-321 which 296 amino acids in length. The C-terminal kinase lobe is composed of one alpha helix and five beta sheets. The N-terminal lobe assist the binding and the coordination of ATP for the transfer of the gamma phosphate to a substrate oriented by the C-terminal lobe. Strands beta-7 and -8 are located within the cleft region between the C-terminal lobe and the N-terminal lobe, they contribute side-chains that involved in catalysis and the binding of magnesium for the coordination of ATP phosphate. Check here to download the monomer form of JNK1-alphal1 isoform of PDB.

FIGURE 3 Structure of KIM docking site within the protein kinase domain shows in pink and ATP binding loop in the cleft shows in green. The Kinase Interaction Motif (KIM) is the docking site for JNK1 to interact with MAPK and it is also called the D-motif or D-sites. It has been found in many MAPK substrats and regulators. [4][6] Mutation of KIM docking site disruptes the ability (up to 95%) of the N-terminal domain of MAPK to bind JNK1. [1] The protein kinase domain must contain a ATP binding site shows in green, otherwise JNK is unable to phosphorylate these protein despite there is a functional JNK docking site.
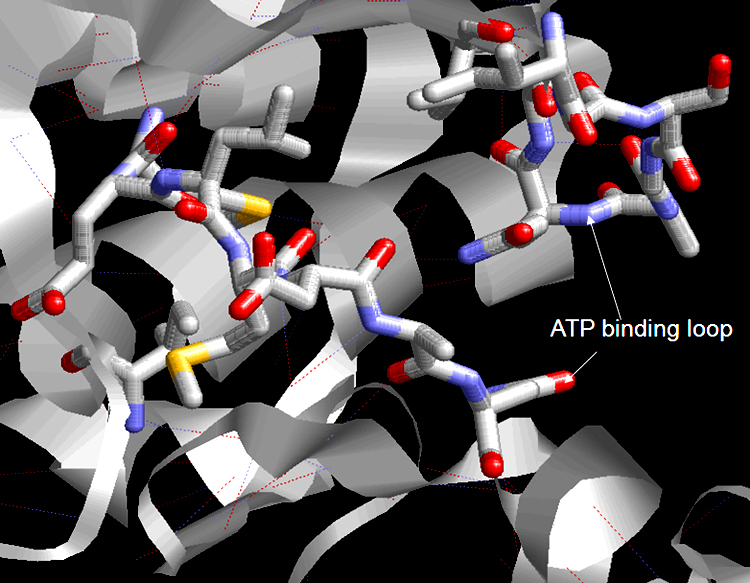
FIGURE 4 Structure of the ATP-binding loop shows in stick form. Amino acid residues 32-40 involved in nucleotide binding of ATP within the ATP binding.

FIGURE 5 Structure of the active site with activation loop shows in yellow. The phosphorylation of threonine 183 and tyrosine 185 within the activation loop, allowing the access of ATP and protein substrates to the active site. Aspartic acid 151 of the DGF motif (amino acid residues 151-153) at the beginning of the activation loop, acts as a proton receptor within the active site and involved ATP binding . [4]
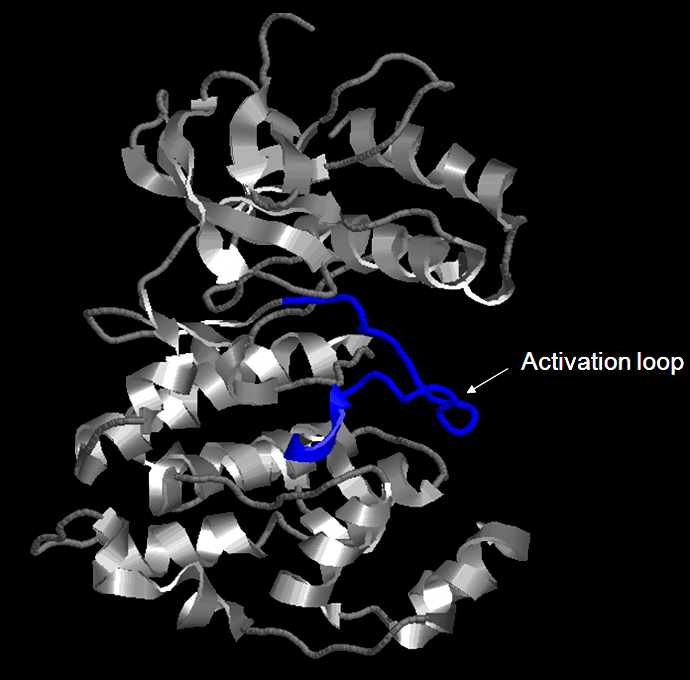
FIGURE 6 Structure of the JNK1-alpha 1 and the activation loop shows in blue. The activation loop (or A loop) is also called the regulatory T-loop and consists amino acid residues 169-190. Upstream protein kinase such as MAPKs can activate JNK by phosphorylating threonine 183 and tyrosine 185 of a conserved TPY motif within the Activation loop. (P45983; Phospho.ELM) The activatation loop undergoes a conformatinal change upon the phosphorylation of both the threonine 183 and tyrosine 185, for full activation of the protein kinases. [3] The phosphorylated tyrosin 185 is the key residue in stabilizing the conformation of the phosphorylated activation loop .
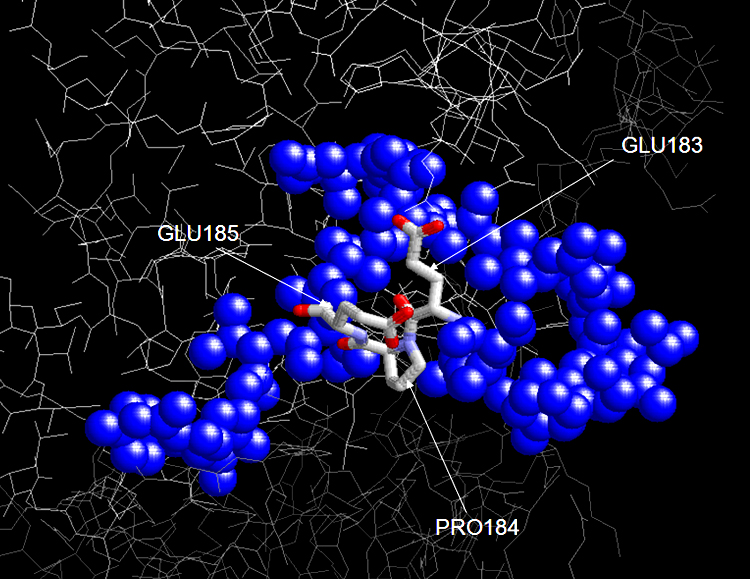
FIGURE 7 Crystal structure of JNK1-alpha1 activation loop with mutations Thr183Glu and Tyr185Glu. The ATP-binding activity of the loop dual specificity for both tyrosine and threonine. Such mutations of the two residues remove the two OH groups and inhibition of phosphorylation is resulted.

FIGURE 8 Structure of alternative amino acids 219-223 in JNK1-alpha1 isoform shows in stick form. There are four Jun N-terminal kinase(JNK)1 isoforms and they show different binding patterns: Beta-1 preferentially binds to c-Jun, whereas alpha-1, alpha-2, and beta- 2 have a similar low affinity of binding to c-Jun. The four isoforms show two different amino acid sequences of residues 219-223: alpha (VCHKI) and beta (IKGGV). However, all four isoforms have the same efficiency for binding and phosphorylation. [2][5]