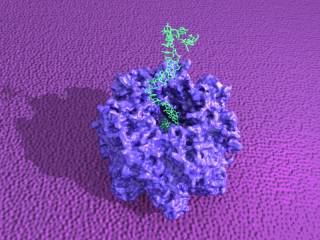
The translocation of polynucleotides through transmembrane protein pores is a fundamental biological process with important technological and medical relevance. The translocation process is complex, and it is influenced by a range of factors including the diameter and inner surface of the pore, the secondary structure of the polymer, and the interactions between the polymer and protein. Computer simulations are a invaluable means of investigating microscopic systems, providing a unique, atomistic perspective of important states and processes.
Our research explores the use of two molecular dynamics simulation translocation methodologies for the nucleotide-nanopore translocation system, with α-hemolysin as the nanopore. The first methodology is the use of non-equilibrium constant velocity-steered molecular dynamics simulations of nucleic acid molecule translocation through the protein nanopore α-hemolysin and the use of Jarzynski's identity to determine the associated free energy profiles. The second methodology is the application of an adaptive biasing force, which can also be used to calculate free energy profiles.
This approach is used to explain the observed differences in experimental translocation time through the nanopore between polyadenosine and polydeoxycytidine. And so we have examined the translocation of polynucleotides and single nucleotides through α-hemolysin. Our simulations provide insight into the role of the interactions between the nucleic acid molecules and the protein pore. We also use mutated protein pores to provide confirmation of residue-specific interactions between nucleotides and the protein pore.
 Close
Close

