WEISS promotes Open Access to research. Any WEISS datasets with no sensitive/identifiable data are hosted here and freely downloadable for evaluation and further research.
| Preview | Description | Web | Ref | Dataset | Size |
|---|---|---|---|---|---|
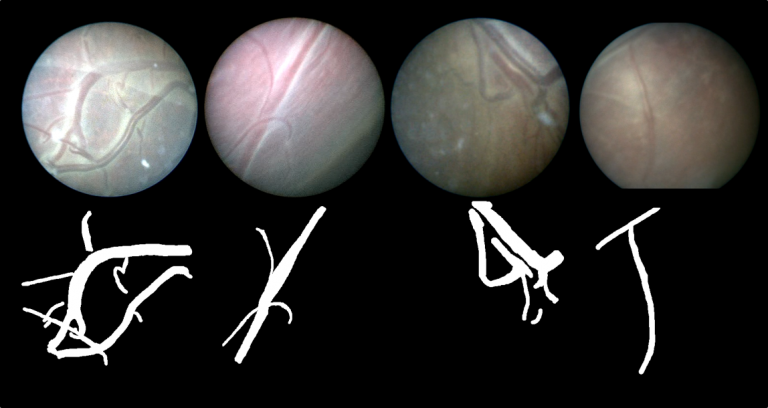 | The dataset contains 483 frames with ground-truth vessel segmentation annotations taken from six different in vivo fetoscopic procedure videos. The dataset also includes six unannotated in vivo continuous fetoscopic video clips (950 frames) with predicted vessel segmentation maps obtained from the leave-one-out cross validation reported in [Bano et al. MICCAI2020]. | URL | [1] | Fetoscopy Placenta Dataset | 450 MB |
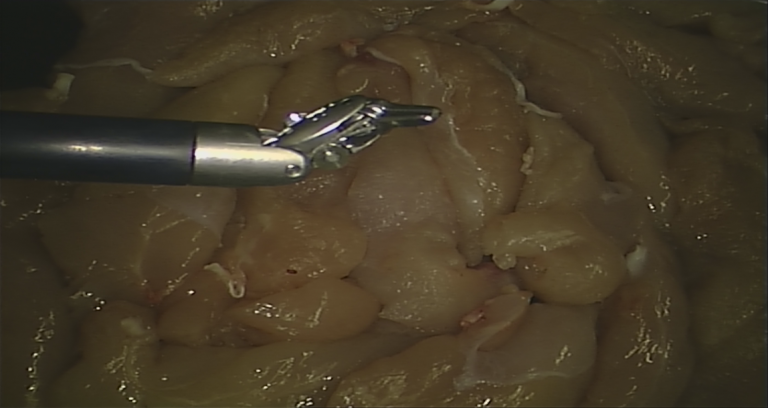 | The dataset consists of 14 + 6 videos of 300 frames each, with corresponding segmentation ground truth and robot kinematic data. Frames were initially recorded at 720x576 pixel resolution and then centrally cropped (538x701) to remove side camera artefacts. The employed technique to produce segmentation labels, as well as further details on our dataset, are accurately described in our [Emanuele et al. MICCAI2020]. | URL | [2] | Synthetic Dataset | 3.5 GB |
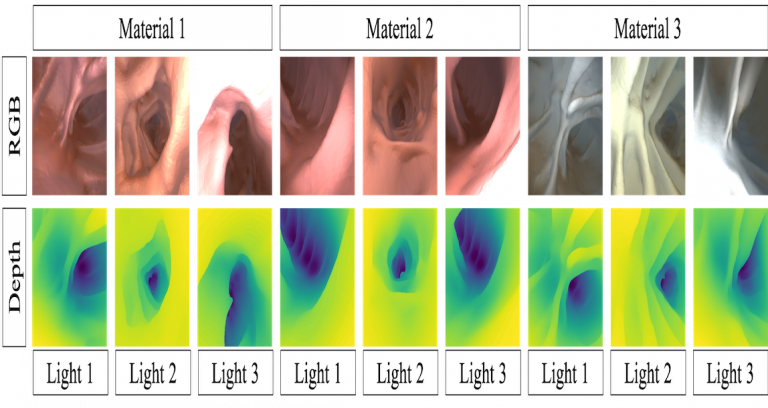 | Synthetic colonoscopy dataset consists of 16,016 RGB images with corresponding ground truth depth. The images are based on a human CTC and were generated in Unity. The depth is scaled to [0,1] which corresponds to [0,20] cm. The data is divided into groups according to its texture (T1, T2, T3) and the lighting (L1, L2, L3). For each configuration there are four to five different subsets generated by randomly shifting and rotating the virtual camera. Please see website and publication for more information. For access permissions, click contact. | URL | [3] | Colonoscopy Dataset | 1.2 GB |
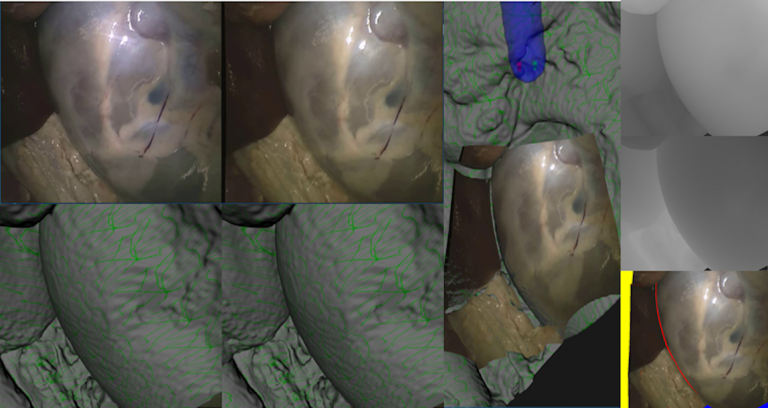 | This dataset consists of 16 rectified stereoendoscopic 720 x 576 resolution images of two porcine ex vivo full torso samples. CT of both endoscope and anatomy enable constrained manual alignment producing reference disparity, depth and occlusion maps for each stereo pair. These are intended for validation of surgical stereo reconstruction algorithms. Reference to follow shortly. | URL | [4] | SERV-CT | 407 MB |
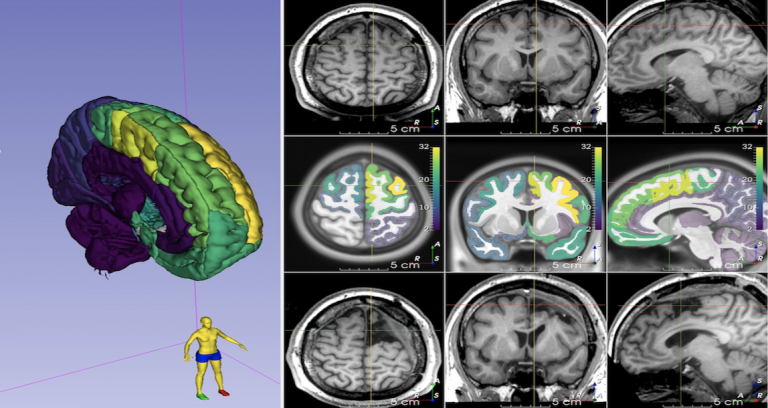 | Descriptive Seizure Semiologies and their multi-one-hot encoded categorical hierarchical brain localisations (11230 datapoints) and lateralisations (2391 datapoints) from 4643 patients across 309 included articles, as a Microsoft Excel Spreadsheet. doi:10.5281/zenodo.4473240 | URL | Semio2Brain | 735 KB | |
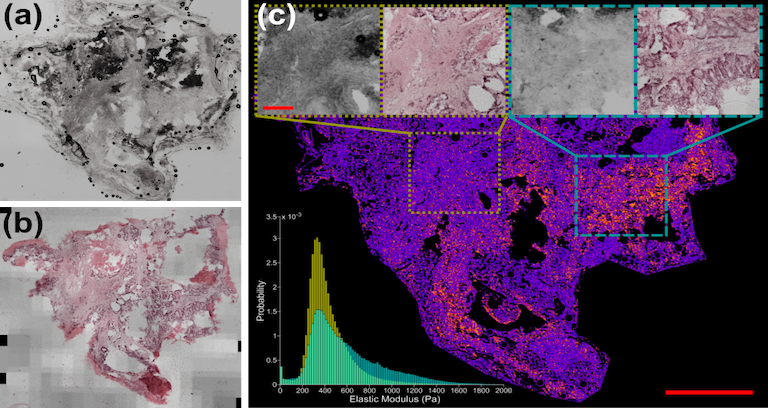 | This dataset consists of 27 tissue samples collected from 7 patients suffering from colorectal or pancreatic cancer metastasis who underwent curative liver resection surgery | URL | AFM Liver Tissue Data | 35 GB | |
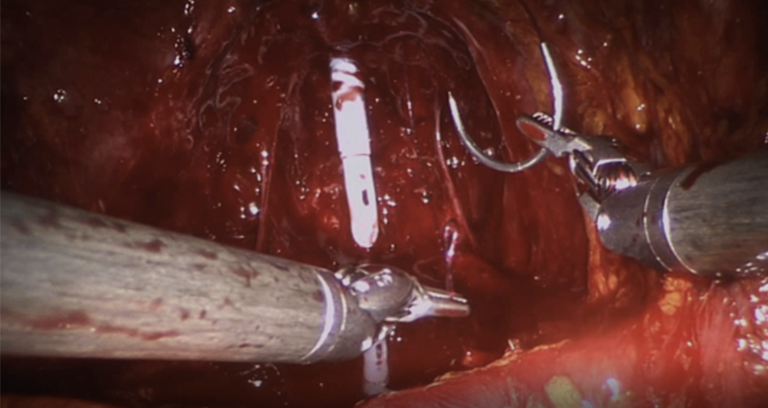 | The dataset contains video segments from 45 Robot-Assisted Radical Prostatectomies (RARP) performed using a da Vinci Si Surgical System (Intuitive Surgical, Inc.). The videos have been manually segmented into 7 fine-grained bimanual gestures and a background class. | URL | [5] | RARP-45 | 24GB |
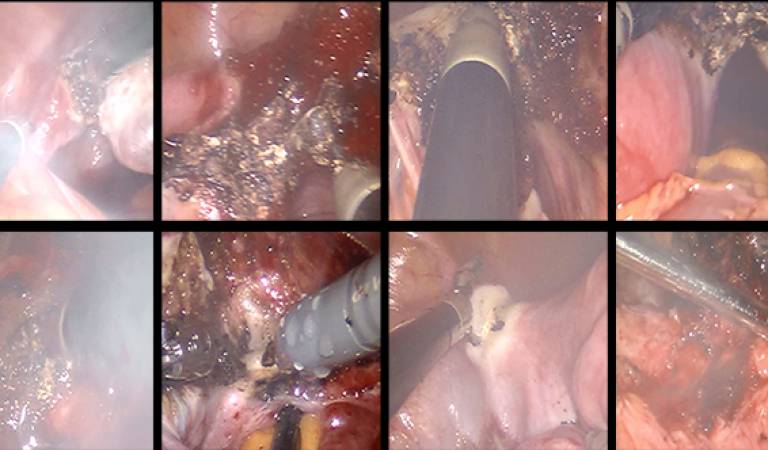 | This dataset contains frames and video clips from 10 robot-assisted laparoscopic hysterectomy procedure videos. The original videos were decomposed into frames at 1 fps. From each video, 300 hazy images and 300 clear images were manually selected by observing the electrocauterisation. A short video clip of 50 frames from each procedure was also selected that was utilised for testing. | URL | [6] | DeSmoke-LAP | 1.8GB |
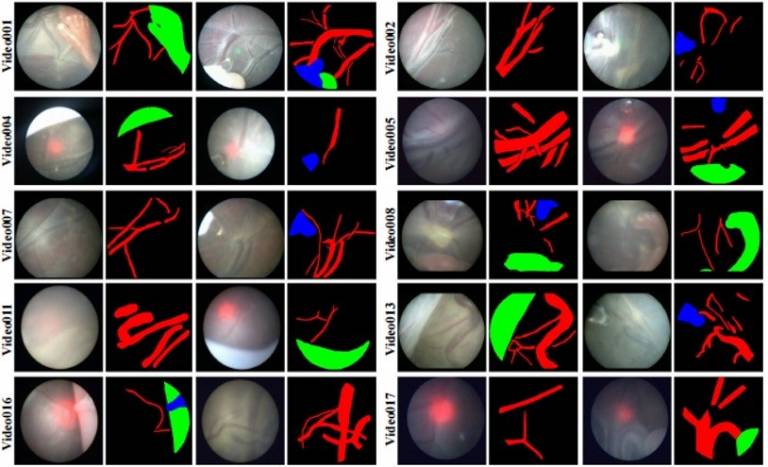 | Fetoscopic Placental Vessel Segmentation and Registration (FetReg2021) challenge was organized as part of the MICCAI2021 Endoscopic Vision challenge. Through FetReg2021 challenge, we released the first large-scale multi-centre dataset of fetoscopy laser photocoagulation procedure. The dataset contains 2,718 pixel-wise annotated images from 24 different in vivo TTTS fetoscopic surgeries and 24 unannotated video clips video clips containing 9,616 frames for training and testing. The dataset is useful for the development of generalized and robust semantic segmentation and video mosaicking algorithms for long duration fetoscopy videos. | URL | [7] | FetReg | 3.2GB |
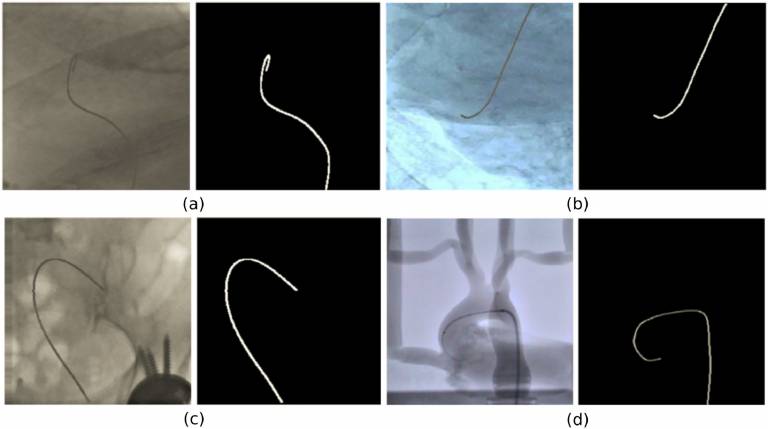 | The dataset contains 3207 images extracted from i) 4 fluoroscopy videos in experiments with an aorta phantom and ii) 6 catheterisation procedures. Binary segmentation masks of the interventional catheter are provided as ground truth. The dataset was used for development and validation of a lightweight U-Net model presented in [Cherardini et. al. 2020]. | URL | [8] | Catheter Segmentation Dataset | 3.51GB |
 | The dataset contains both instrument and surgical skill assessment annotations in a high-fidelity bench-top phantom of the nasal phase of endoscopic pituitary surgery (15-videos). | URL | Pituitary Surgery UpSurgeon Dataset | 9.06GB | |
 | The dataset contains both surgical step and surgical instruments annotations for 25-videos of endoscopic pituitary surgery. | URL | PitVis-2023 Workflow Recognition Dataset | 39.87GB |
References:
- Bano, S., Vasconcelos, F., Shepherd, L.M., Vander Poorten, E., Vercauteren, T., Ourselin, S., David, A.L., Deprest, J. and Stoyanov, D., 2020, October. Deep placental vessel segmentation for fetoscopic mosaicking. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 763-773). Springer, Cham.
- Colleoni, E., Edwards, P. and Stoyanov, D., 2020, October. Synthetic and Real Inputs for Tool Segmentation in Robotic Surgery. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 700-710). Springer, Cham.
- Rau, A., Edwards, P.E., Ahmad, O.F., Riordan, P., Janatka, M., Lovat, L.B. and Stoyanov, D., 2019. Implicit domain adaptation with conditional generative adversarial networks for depth prediction in endoscopy. International journal of computer assisted radiology and surgery, 14(7), pp.1167-1176.
- Edwards, P.J., Psychogyios, D., Speidel, S., Maier-Hein, L. and Stoyanov, D., 2020. SERV-CT: A disparity dataset from CT for validation of endoscopic 3D reconstruction. arXiv preprint arXiv:2012.11779.
- Van Amsterdam B, Funke I, Edwards E, Speidel S, Collins J, Sridhar A, Kelly J, Clarkson MJ, Stoyanov D., 2022. Gesture Recognition in Robotic Surgery with Multimodal Attention. IEEE Transactions on Medical Imaging.
- Pan, Yirou and Bano, Sophia and Vasconcelos, Francisco and Park, Hyun and Jeong, Taikyeong Ted. and Stoyanov, Danail, 2022. DeSmoke-LAP: Improved unpaired image-to-image translation for desmoking in laparoscopic surgery. International Journal of Computer Assisted Radiology and Surgery.
- Bano, S., Casella, A., Vasconcelos, F., Qayyum, A., Benzinou, A., Mazher, M., Meriaudeau, F., Lena, C., Cintorrino, I.A., De Paolis, G.R. and Biagioli, J., 2022. FetReg2021: A Challenge on Placental Vessel Segmentation and Registration in Fetoscopy. arXiv preprint arXiv:2206.12512.
Marta Gherardini, Evangelos Mazomenos, Arianna Menciassi, Danail Stoyanov, “Catheter segmentation in X-ray fluoroscopy using synthetic data and transfer learning with light U-nets”, Computer Methods and Programs in Biomedicine, Volume 192, Aug 2020, 105420, doi:10.1016/j.cmpb.2020.105420.
 Close
Close

