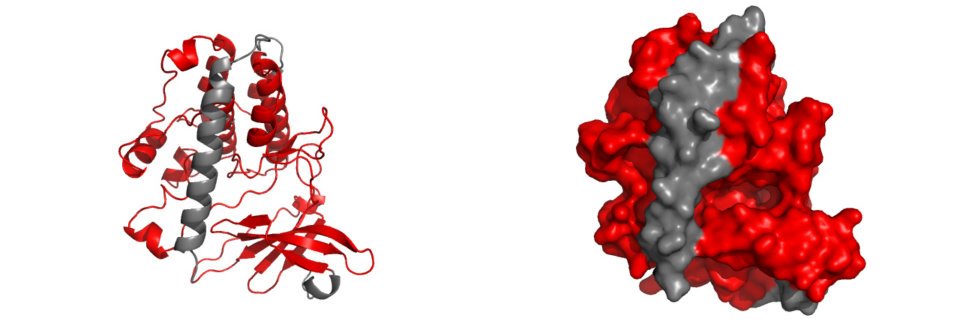
The entire catalytic domain of CaMKIV is displayed in the diagram above, represented by the red highlighted structures. Upon closer inspection, it is evident that the catalytic domain is infact subdivided into many smaller structures. A breakdown of these structures is displayed below, which includes ATP binding residues, C-helix, Cataltic loop, Activation loop and Substrate binding residues. Each of these domains play a critical role in the behaviour of CaMKIV following different signal stimulus, and collectively, they allow for the correct and regulated function of CaMKIV in the CaMKIV PATHWAY. |
| The catalytic domain is shown in bold in all sequences |
ATP binding residues |
There are different residues involved in ATP binding once CaMKIV has been activated. These are: - Glycine rich loop residues (red). The three residues backbone amines form H bonds to the ATP β phosphate to orientate it in a ‘top’ position, where the adenine ring is at the outer space of the ATP binding site. - Valine 60 (blue) from β2 strand forms a hydrophobic interaction with the adenine ring of ATP. - Lysine 75 (yellow) from the β3 strand coordinates the non-hydrolysable phosphates of ATP laterally (i.e. α and β phosphates), and is thus heavily involved in the coordination of the way in which ATP binds to CaMKIV. - Proton accepting residue, D 164 (orange), is the catalytic base of the reaction, abstracting a proton from a target –OH group of the substrate protein. The resulting aloholate or phenolate ion is optimally positioned to attack the γ phosphate of ATP. - Lysine 166 (cyan) is probably the most important residue in the ATP binding site, given its capability of recognition of γ phosphate and hence, its ability of differentiating between ATP and ADP. A few highly conserved residues on each side of it help in the positioning of the phosphoacceptor residue of the peptide substrate. - Highly conserved DFG triplet (magenta) coordinates 2 Mg +2 ions, which in turn coordinate to ATP α and β phosphates, from a bottom position. |
C-helix |
The C-helix contains the conserved glutamic acid, E, through a short loop. E 89 interacts via salt bridge interaction to K 75 (ATP binding site) to directly coordinate α and β phosphates of ATP. |
Catalytic loop |
The catalytic loop is located towards the N-terminus section of CaMKIV. It is key in the phosphotransfer mechanism and in orientating peptide substrates to the active site. D 164 and K 166 are in the catalytic loop and were previously exposed in the ATP binding sites. The C-terminal portion of the loop (TEN) is a 3 10 helix which is more tightly wound than an α helix. |
Activation loop |
The activation loops of CaMKIV are located within the catalytic domain and are shown here in olive green. In its active state, the activation loop adopts an extended conformation that sprawls laterally away from the active site along the N lobe before redirecting back to the active site at the C lobe. Right orientation of this loop plays an important role in orientating the peptide substrate for phosphotransfer through H bonds. Thr 200, regulatory residue, is autophosphorylated upon activation. In its inactive state, the loop is really mobile, mostly due to the lack of ATP in the catalytic site, which promotes locking of the loop in its active sate. This mobility of the loop in is reflected in the X-ray crystallography since the loops are invisible in the structure found on PDB but are seen in this picture since a modelling program has been used (SwissModel). |
Substrate binding residues |
Shown in the picture above is the substrate binding site in cyan. The autoinhibitory domain and how its removal by a conformational change removes inhibition is clearly visible. There are three residues which undergo phosphorylation in order to regulate the activity of CaMKIV. Thr 200 is one of the residues, and Ser 12 and Ser 13 are the other two. There are also three steps involved for CaMKIV to be activated. These are: 1) The binding of Ca 2+/calmodulin to CaMKIV 2) Phosphorylation of the Ca 2+/calmodulin-bound enzyme 3) Autophosphorylation of Ser 12-Ser 13.(These residues are not shown since the model starts at amino acid 34) |
References |
1. ATP binding residues: (Chatila T., K.A. Anderson, N. Ho and A.R. Means. 1996. A Unique Phosphorylation-dependent Mechanism for the Activation of Ca2+/Calmodulin-dependent Protein Kinase Type IV/GR. JBC271:21542-21548) (Cesareni, Gimona, Sudol and Yaffe (Eds), Modular Protein Domains, 2004) 2. C-helix: (Green M.F., K. Anderson, A.R. Means. 2011. Charaterization of the CaMKKβ-AMPK signaling complex. Cel. Sig.23:2005-2012) 3. Substrate binding residues: (Chatila T., K.A. Anderson, N. Ho and A.R. Means. 1996. A Unique Phosphorylation-dependent Mechanism for the Activation of Ca2+/Calmodulin-dependent Protein Kinase Type IV/GR*. JBC271:21542-21548) |