Multiple Sequence Alignment
We have performed a first multiple sequence alignment of the four human
isoforms, followed by a multiple sequence alignment of the human isoform
a (which is the most abundant isoform in humans), the LAT homologue
in rat, the LAT homologue in mouse and the LAT homologue in cow.
This was done using the Clustal W tool provided by the Swiss Institute
of Bioinformatics , followed by BOXSHADE 3.21 which is a printing and
shading of multiple alignment files.
Colour code:
Black: conserved residues
Grey: conservative mutations
White: divergence
According to the predicted linear model presented here, we have indicated
the possible domains interacting with PLC- γ1 and with GRB2, GRAP2
and PIK3R1. We have also indicated the extracellular topological domain
and transmembrane region of the human isoforms
MULTIPLE SEQUENCE ALIGNMENT OF THE HUMAN LAT ISOFORMS

From analysing four isoforms of LAT our MSA shows large sections of
the protein are conserved across all forms (shown by the amino acids
highlighted in black). The amino acids that have been identified as
key sites for signal transduction are preserved in all four isoforms
indicating that they are able to carry out the same functions. Isoform
A is the most common form of LAT protein found so for the next multiple
sequence alignment we have just compared to this.
MULTIPLE SEQUENCE ALIGNMENT
OF THE LAT HOMOLOGUES IN DIFFERENT SPECIES
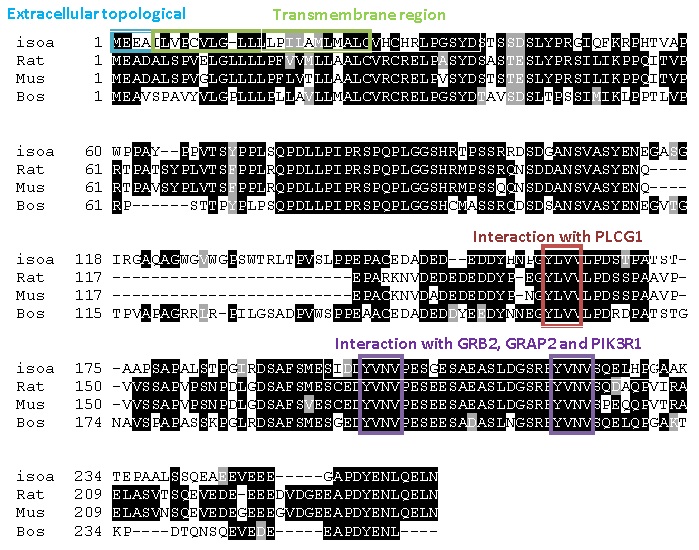
From this MSA you can see there are many sections of the protein that
are conserved across all four species, shown by the amino acids highlighted
black. There are also individual amino acids highlighted grey showing
that the sequence contains many amino acids with similar characteristics/properties
to those in the same position on the different forms of LAT. This is
important for binding to ensure the same regions always have distinct
properties so can be recognised by their target substrates, for example
residues with the binding site could be hydrophobic, negatively charged
etc..
Using the predicted linear model we have produced, we have indicated
the possible domains interacting with PLCγ1 , GRB2, GRAP2 and PIK3R1.
These domains are conserved across all the alignments showing all the
different forms of LAT bind to their target in the same regions. GRB2,
GRAP2 and PIK3R1 all bind via their SH2 domain to the same region on
the LAT protein [1]; this shows these proteins under certain conditions
all recognise the same consensus sequence. SH2 domains recognise and
bind to tyrosine phosphorylated sites. Looking at the highlighted interaction
domains the tyrosine residue is always followed by Valine and Asparagine,
hydrophobic and polar residues. We know the SH2 domain on GRB2 preferably
binds to Y-X-N-X [2], where X is a hydrophobic residue (eg.Valine),
which fits perfectly with all these alignments.
PLC γ1 also interacts with phosphorylated tyrosine residues [1] by cleaving
the phospholipid. From the interaction domain for PLC γ1 the tyrosine
residue this time is followed by Valine and Leucine residues, which
are both hydrophobic.
The first four residues of LAT isoform a are the extracellular domain
which are hydrophobic or negatively charged this is followed by the
membrane spanning region (23 residues); these are mainly hydrophobic
as well.
The differences between any of the different forms of LAT could be
due to alternative splicing; rearrangement of RNA exons produced during
transcription of LAT gene. Both the LAT form found in mice and rats
show huge sections that are missing compared to the Human isoform a
and LAT found in cows, this could be due to a deletion due to the information
coded not being required in the rat or mouse. A – indicates missing
parts of the gene compared to other alignments.
References
[1]. Wange R.L, (2000). “LAT, the linker for activation of T cells: a bridge between T cell-specific and general signaling pathways”. Science's STKE : signal transduction knowledge environment, Vol.2000(63), p.re1.
[2]. Nioche P, (2002). “Crystal structures of the SH2 domain of grb2: highlight on the binding of a new high-affinity inhibitor”. Journal of Molecular Biology, Vol.315(5), p.1167-1177.